|
|||||
|
|||||||
|
    An initiative of :Stichting Food-Info
|
| Food-Info.net> Topics > Food Safety > Food-borne viruses An introduction to viral classification and replicationAdapted from http://www.tulane.edu/~dmsander/WWW/224/Classification224.html ClassificationClassification makes possible predictions about details of replication, pathogenesis and transmission. This is particularly important when a new virus is identified. It also means that when a new species of known virus family or genus is investigated it can be done in the context of the information that is available for other members of that group. Without a classification scheme each newly discovered virus would be like a black box. Nothing could be assumed, everything would have to be discovered and rediscovered. These reasons dictate that the development of a classification scheme is important and inevitable. The current classification scheme allows most newly described viruses to be placed in a box with a label. In the best cases much can be assumed about the biology of the virus. Even in the worse case a framework for investigation would be suggested. Because there are so few virus discoveries now being made which do not fit into the existing classification scheme we can state with a degree of confidence that most of the major groupings of viruses infecting humans and domesticated animals have been identified. So classification is useful. How are viruses classified ?Hierarchical virus classification: order - family - subfamily - genus - species - strain/type At the moment classification is really only important from the level of families down. All families have the suffix - viridae e.g.
Members of the family Picornaviridae are generally transmitted via the faecal/oral and airborne routes. Genera have the suffix - virus. Within for example the Picornaviridae there are 5 genera:
The definition of `species' is the most important but difficult assignment to make with viruses. There is an element of subjectivity about it. Consider the Lentivirus genus, it is known to contain many different species including the following:
But there are viruses that are intermediate between two described species. Should these be a different species or included with a known species ? Such variations are mainly a feature of viruses with an RNA genome and cause a lot of difficulties in classification. Basis of Taxonomic Classification.Features such as morphology (size, shape, enveloped/unenveloped), physicochemical properties (molecular mass, buoyant density, pH, thermal, ionic stability), genome (RNA, DNA, segmented sequence, restriction map, modifications etc), macromolecules (protein composition and function), antigenic properties, biological properties (host range, transmission tropism etc) are all considered. The Baltimore ClassificationThe replication strategy of the virus depends on the nature of its genome. Viruses can be classified into seven (arbitrary) groups: 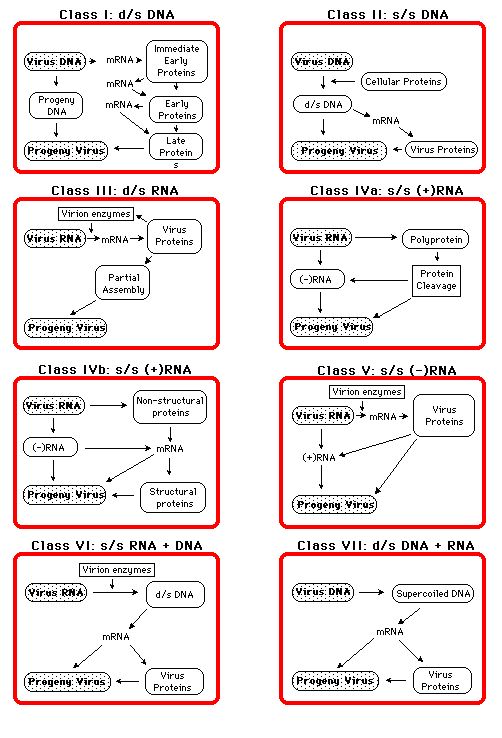
ReplicationFor a virus to multiply it must obviously infect a cell. Viruses usually have a restricted host range i.e. animal and cell type in which this is possible. All must make proteins with 3 sets of functions. - ensure replication of the genome - package the genome into virus particles - alter the metabolism of the infected cell so that viruses are produced. 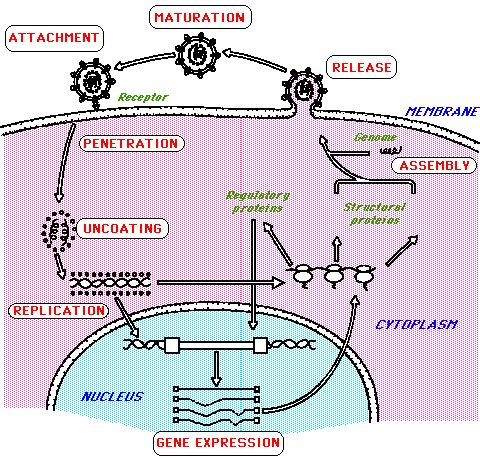 This actually shows the replication of a retrovirus, HIV-1 but the main events are common to all viruses although the precise details obviously vary. Initiation phase:
The genetic material of the virus is introduced into the cell, often accompanied by essential viral protein cofactors. Replication phase:
The genomic size, composition and organization of viruses shows tremendous diversity (look at the above Baltimore classification). In a few instances it is cellular enzymes that replicate the viral genome, assisted by viral proteins e.g parvovirus. In most cases the opposite is true, viral proteins are responsible for genome replication although they utilise cellular proteins to aid this. Release phase:
In all instances it is viral proteins that are responsible for this third and final stage although host proteins and other factors may still associate with the virus particle. Particle/infectivity ratio in picornaviruses is as low as 0.1% i.e. only 1 in 1000 virion particles are infectious. A virus can be defective for innumerable reasons but it makes the study of replication difficult. More reading on viruses : http://www.virology.net/ |
|
| ||
| Food-Info.net is an initiative of Stichting Food-Info, The Netherlands | ||||||